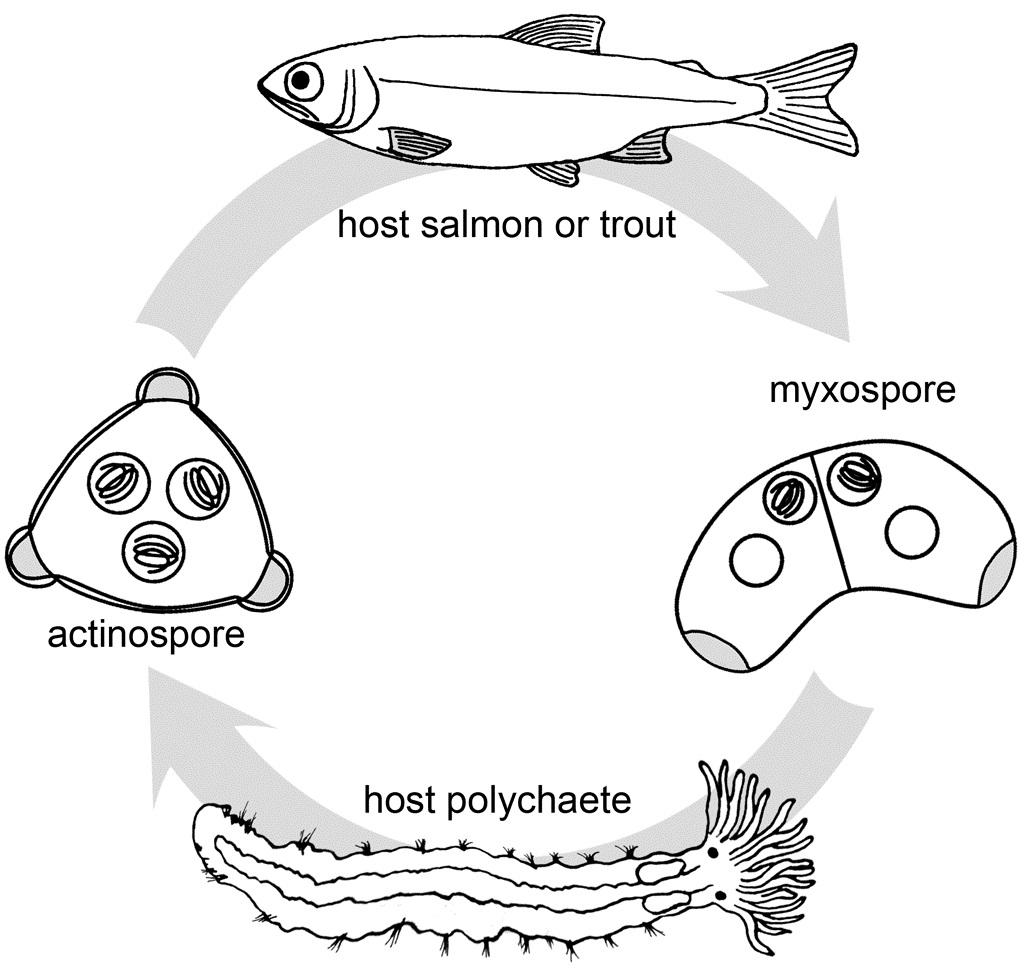
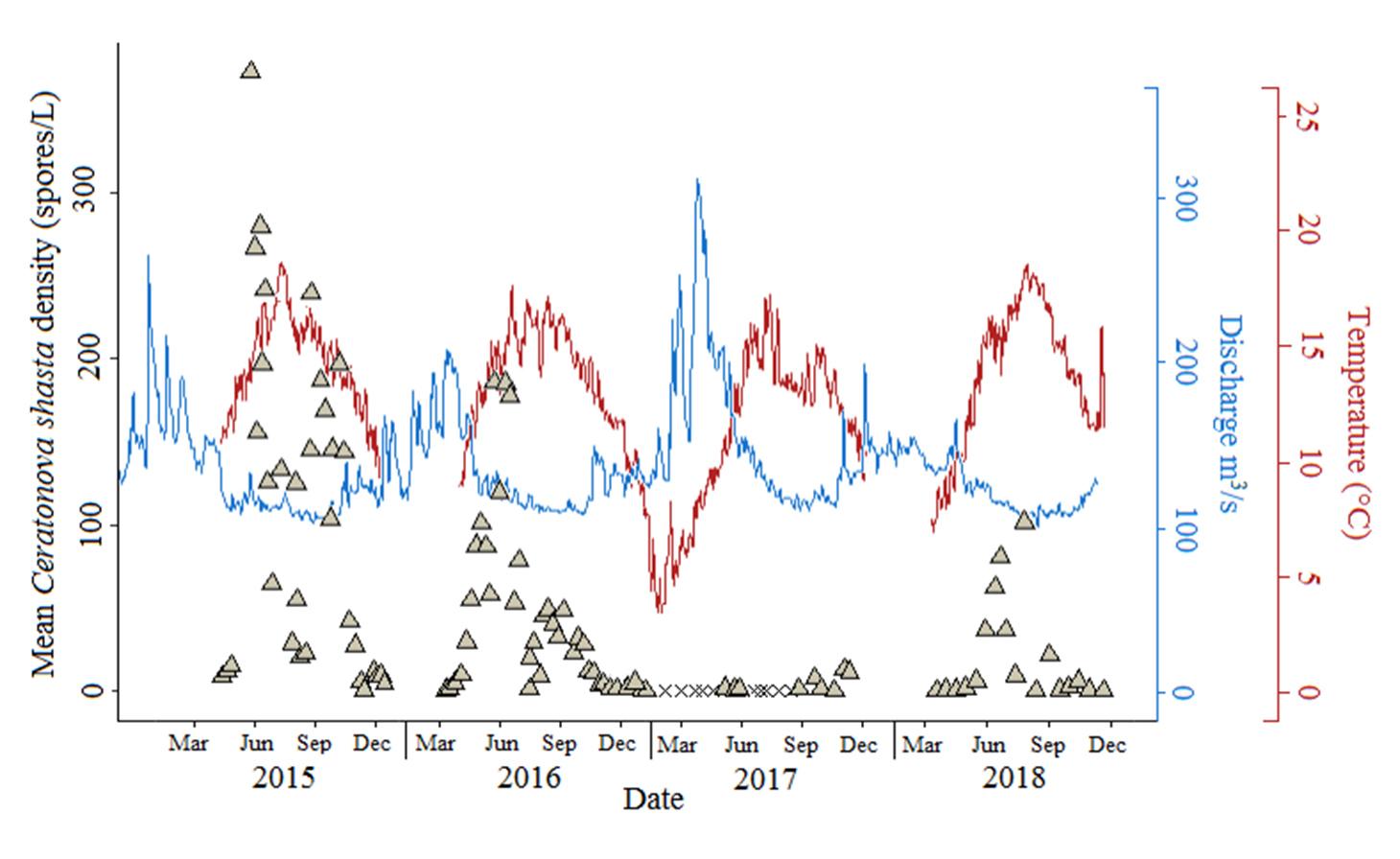
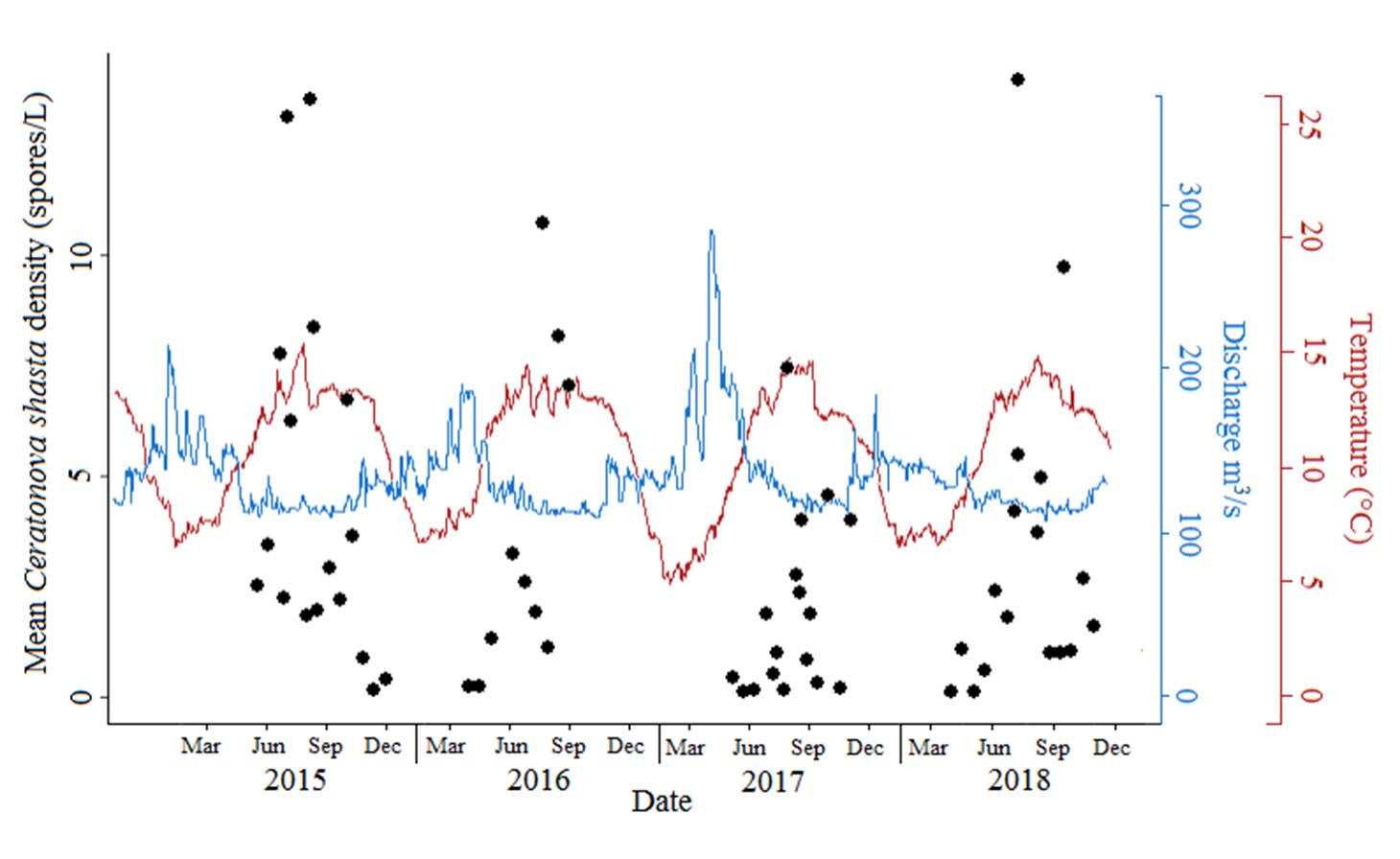
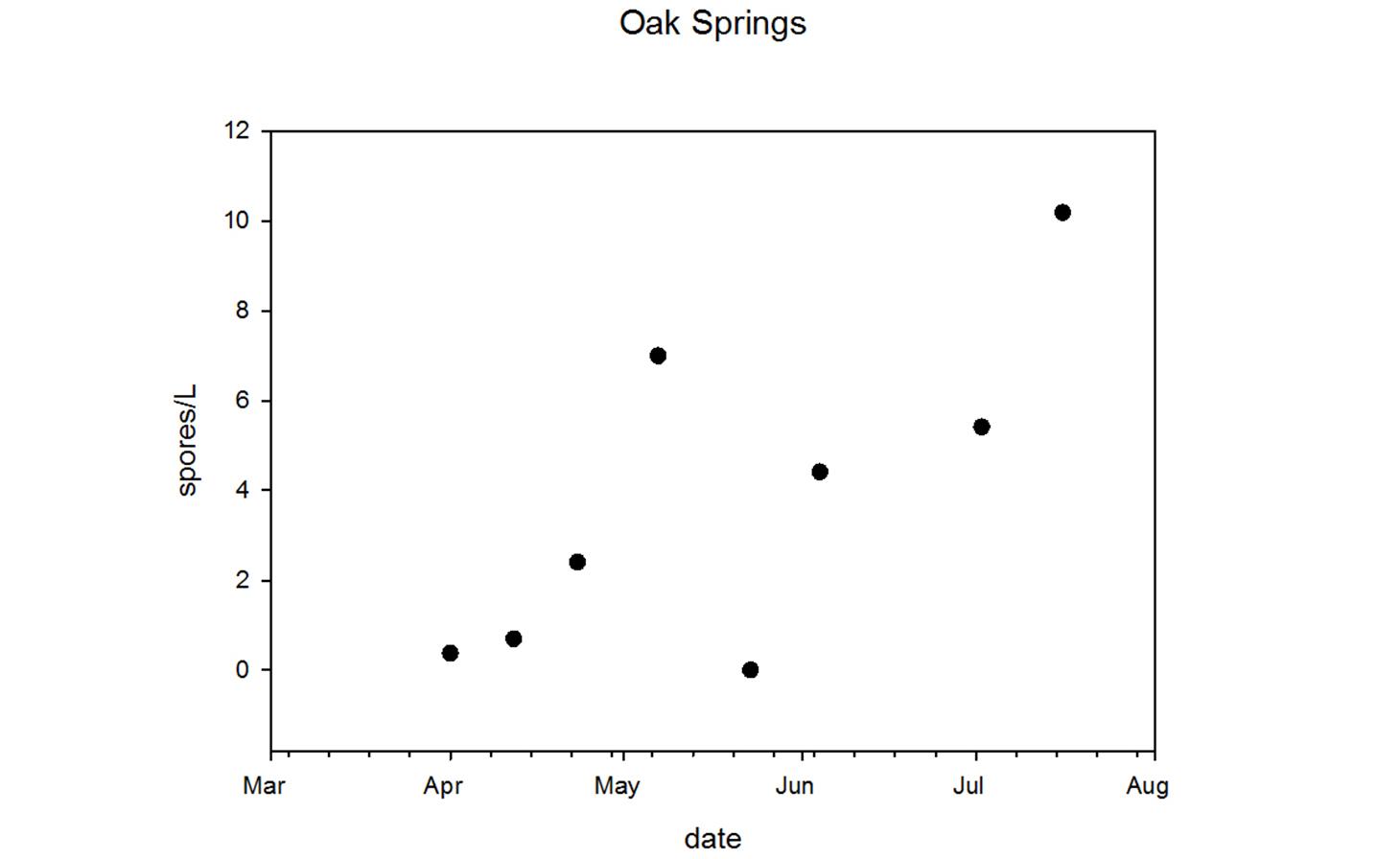
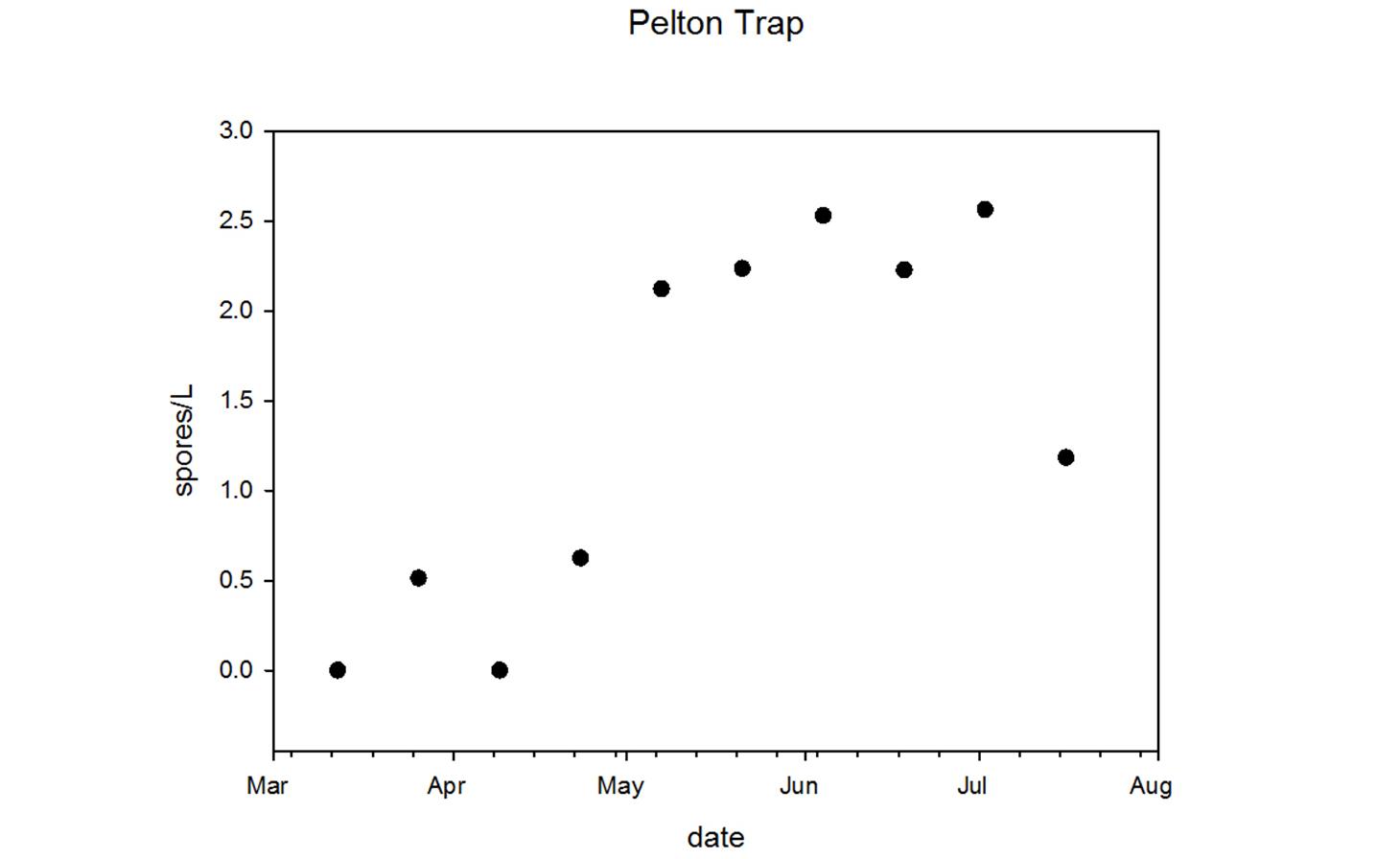
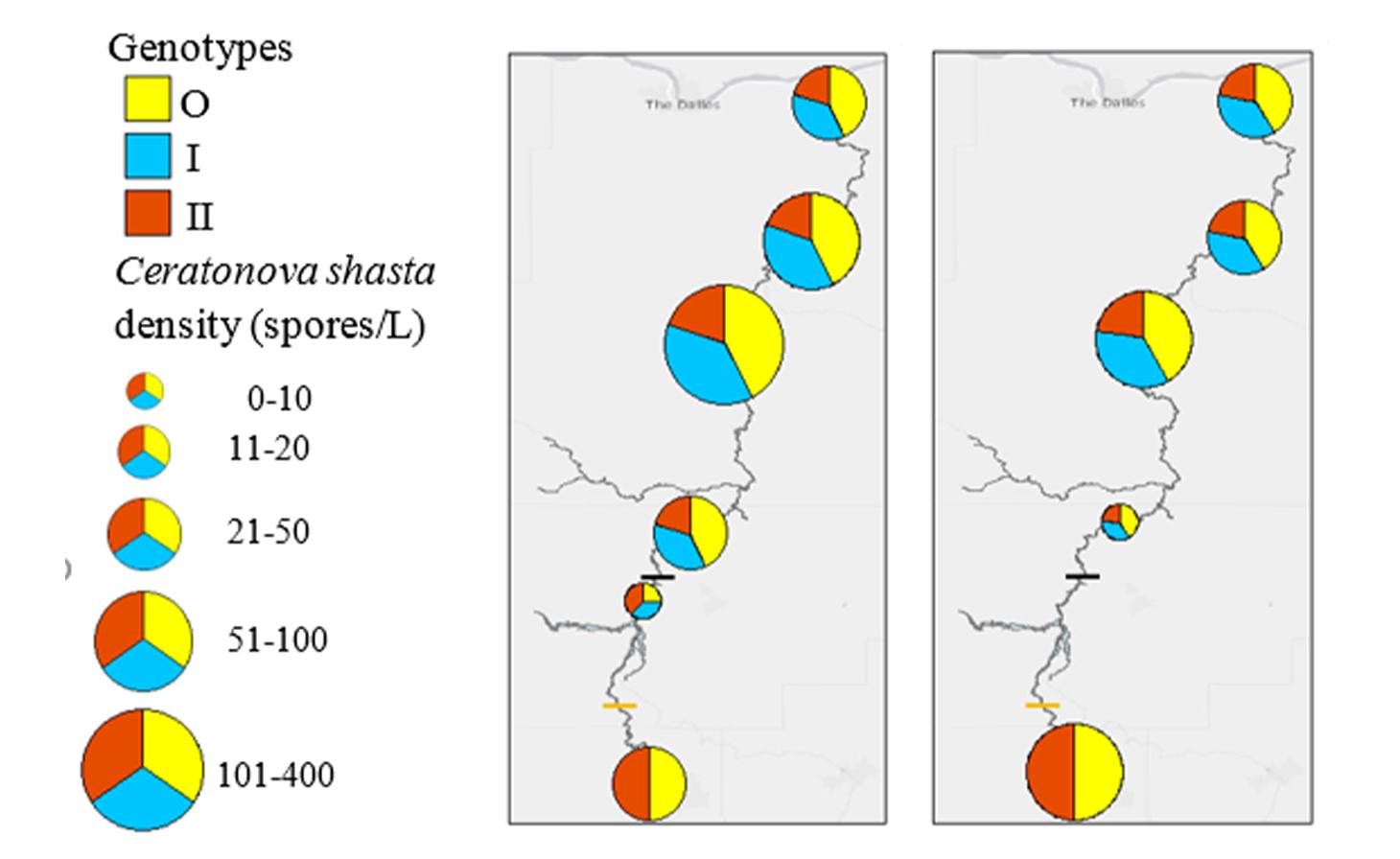
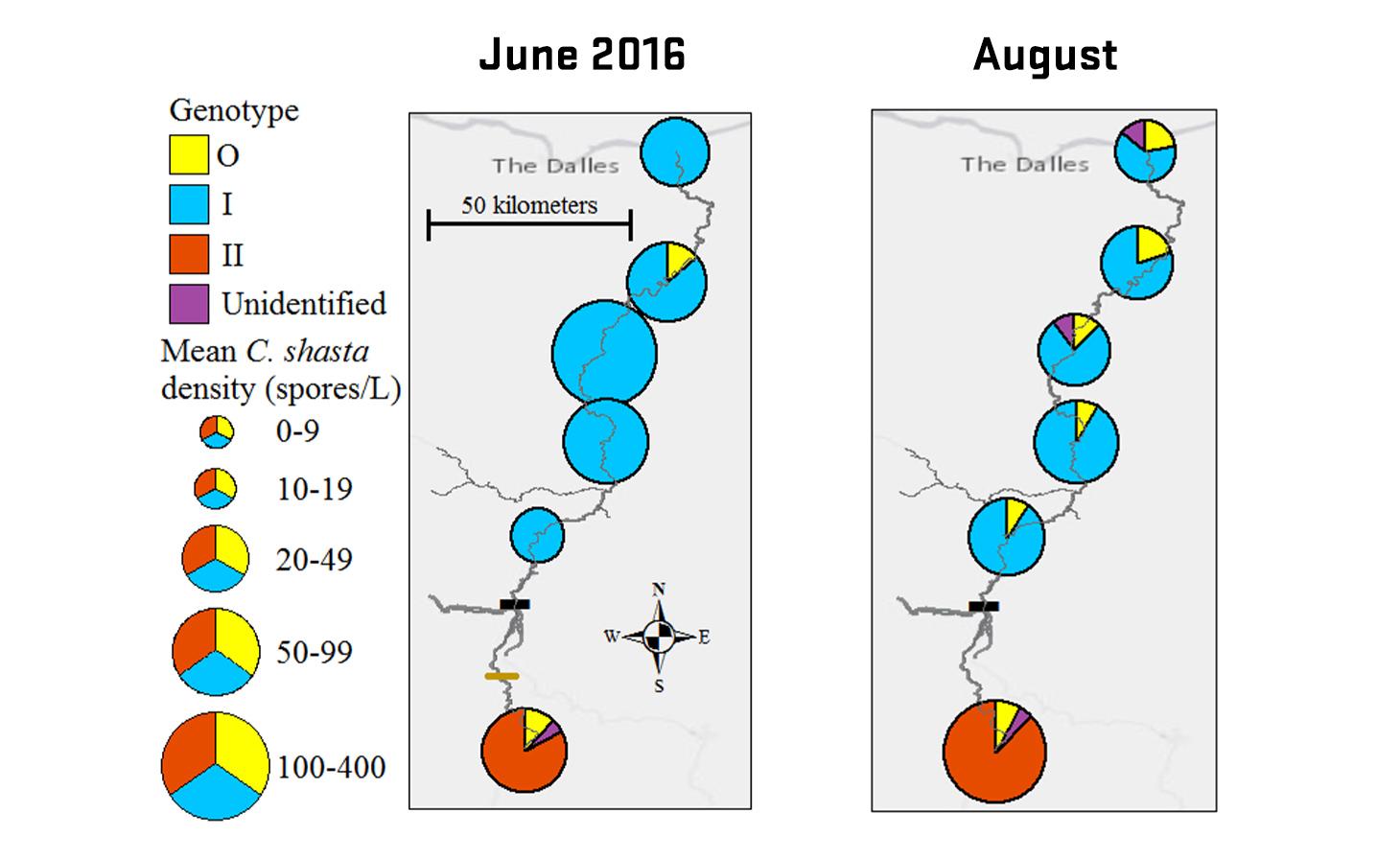
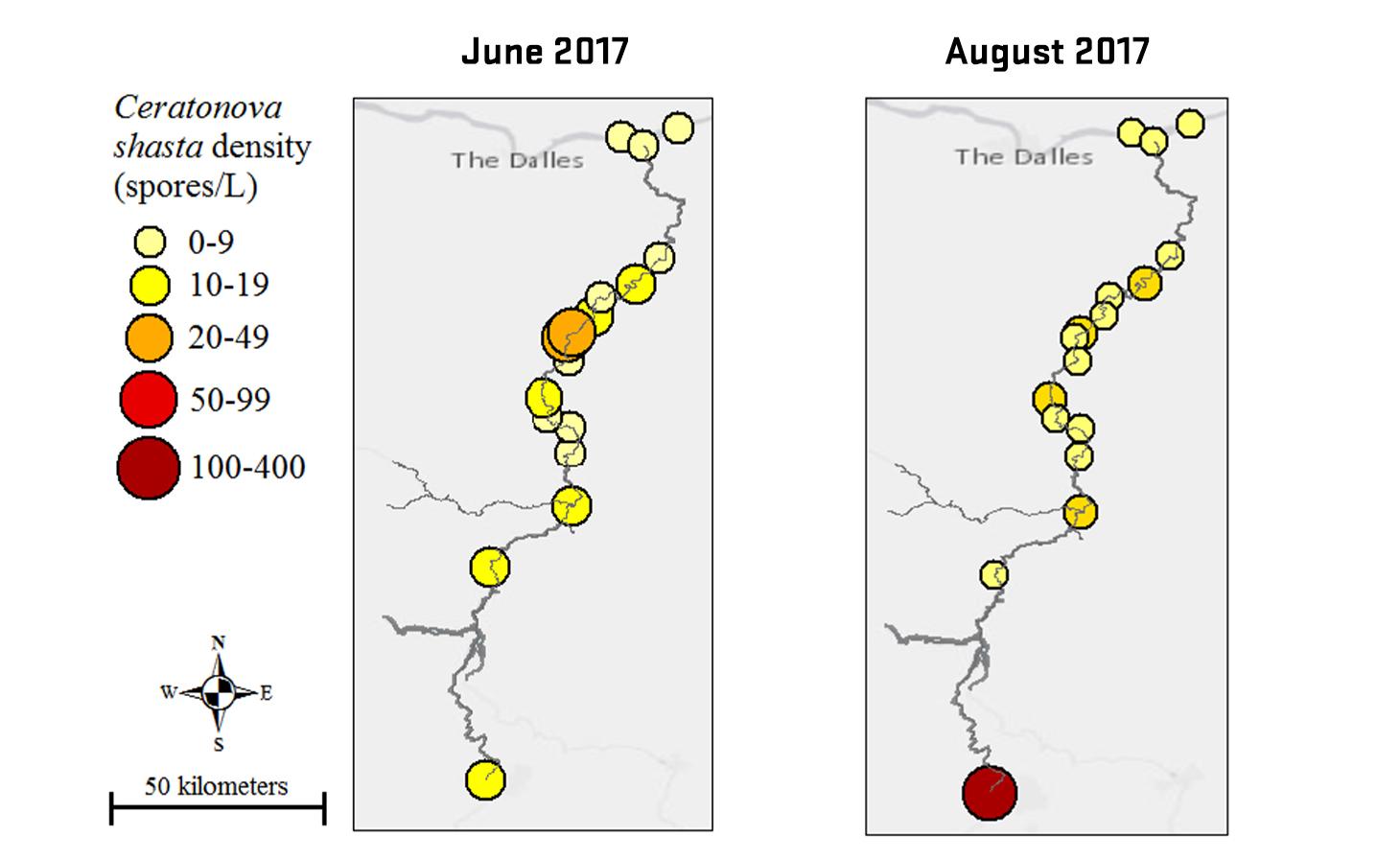
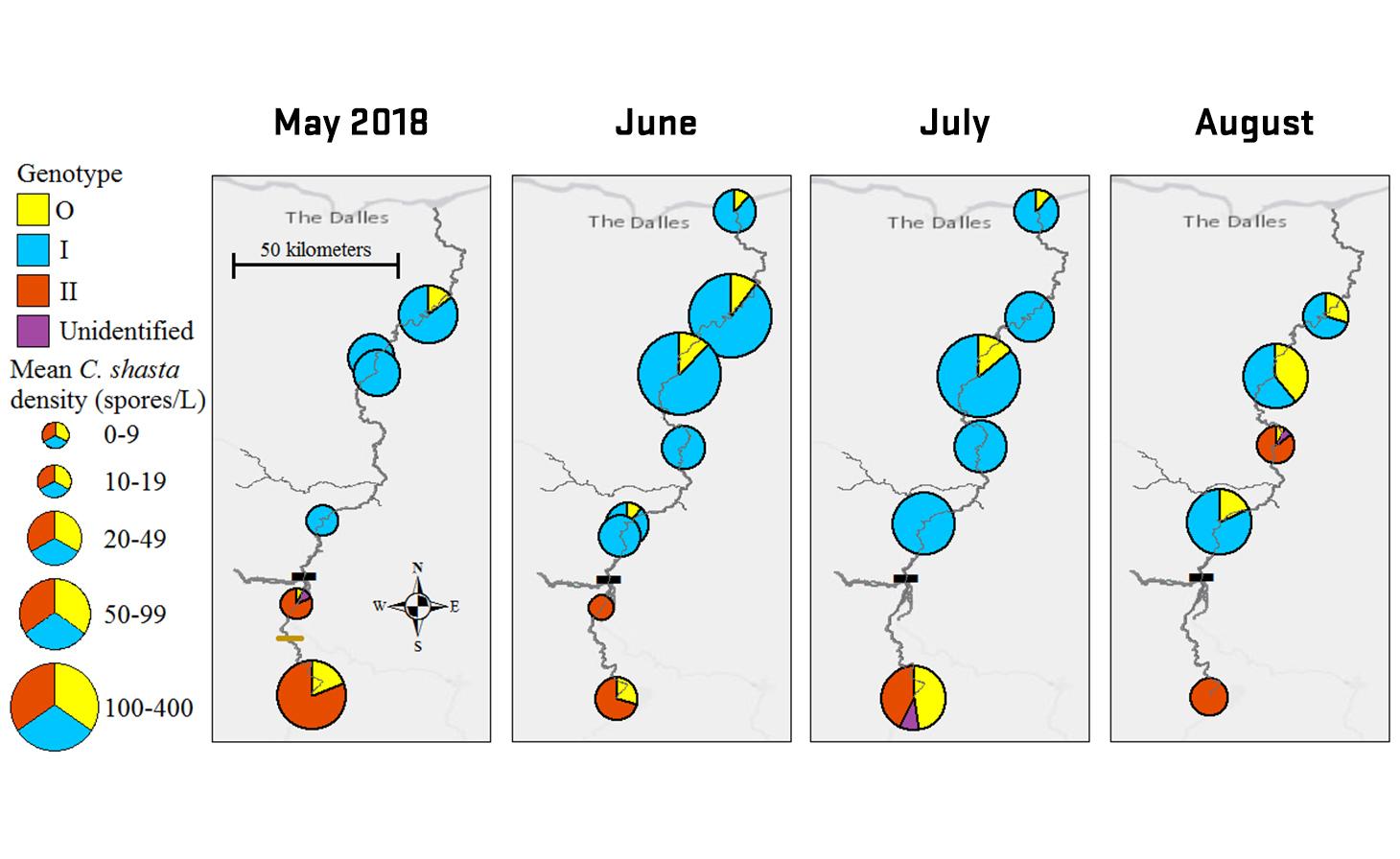
Ceratonova shasta is a freshwater, myxozoan parasite that is native to the Pacific North West of North America. It causes enteronecrosis in salmonids which, in the Deschutes River Basin, has resulted in high mortality in cultured juvenile salmon at the Pelton Facility and in adult salmon returning to both the Round Butte and Warm Springs hatcheries. Transmission occurs through waterborne stages: actinospores released from polychaete worms infect salmonid fishes and develop into myxospores which then infect polychaetes (see life cycle on left). The parasite proliferates in each host (not in water).
“By understanding the relationships between the parasite, its hosts, and the natural habitats and conditions that are favorable or unfavorable for infection, strategies can be developed to limit the exposure of fish to areas where the risk of infection of C. shasta may be higher, or develop management strategies based upon predicted water and climate conditions to improve the survival and viability of wild and hatchery salmonids.” Cyndi Baker, PhD, CTWSRO.
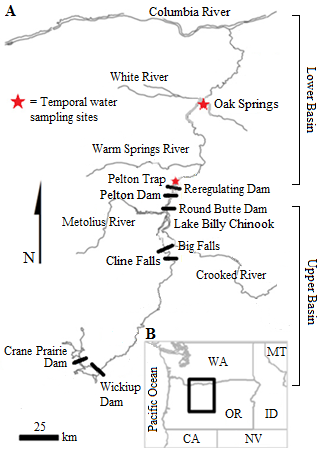
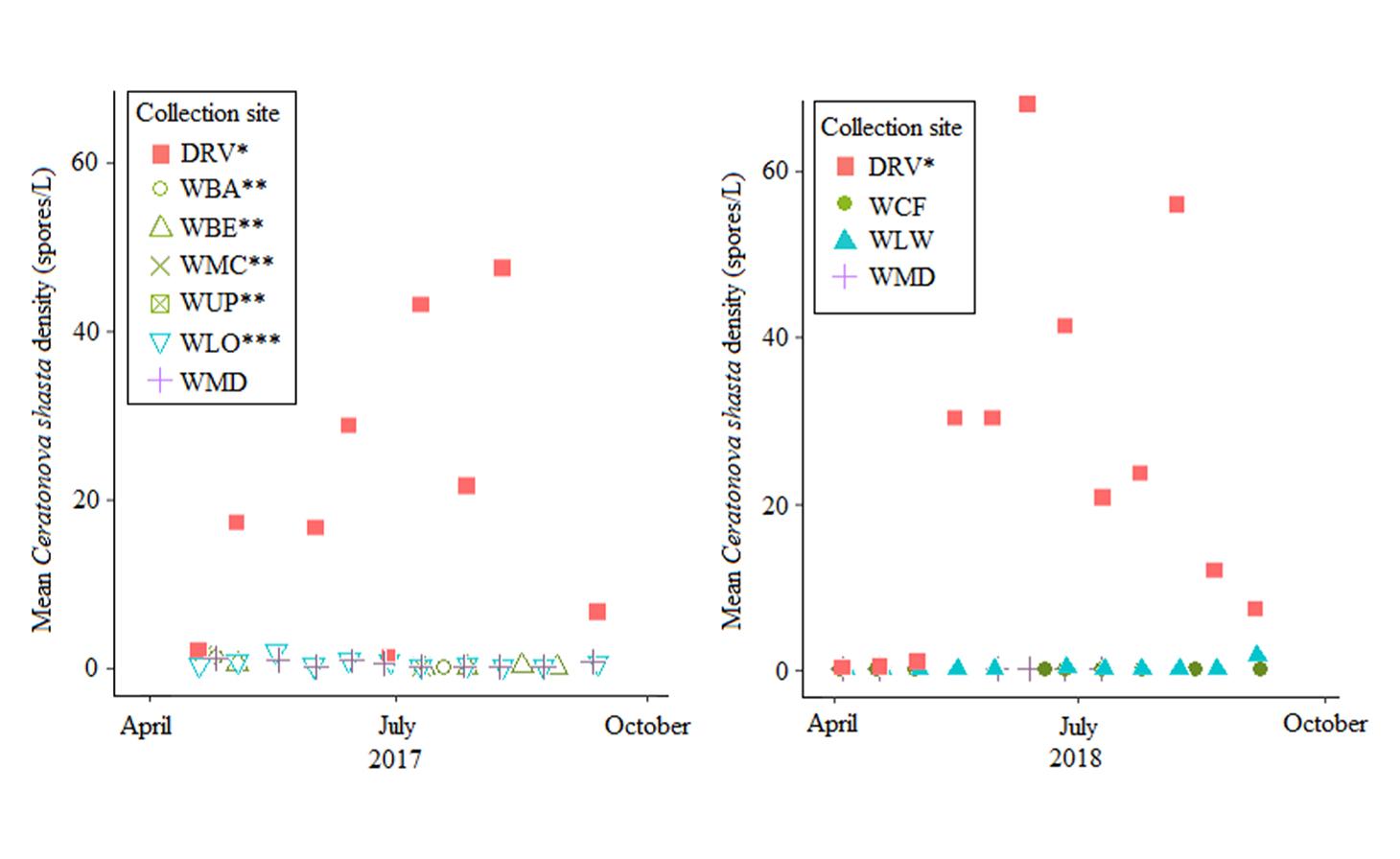
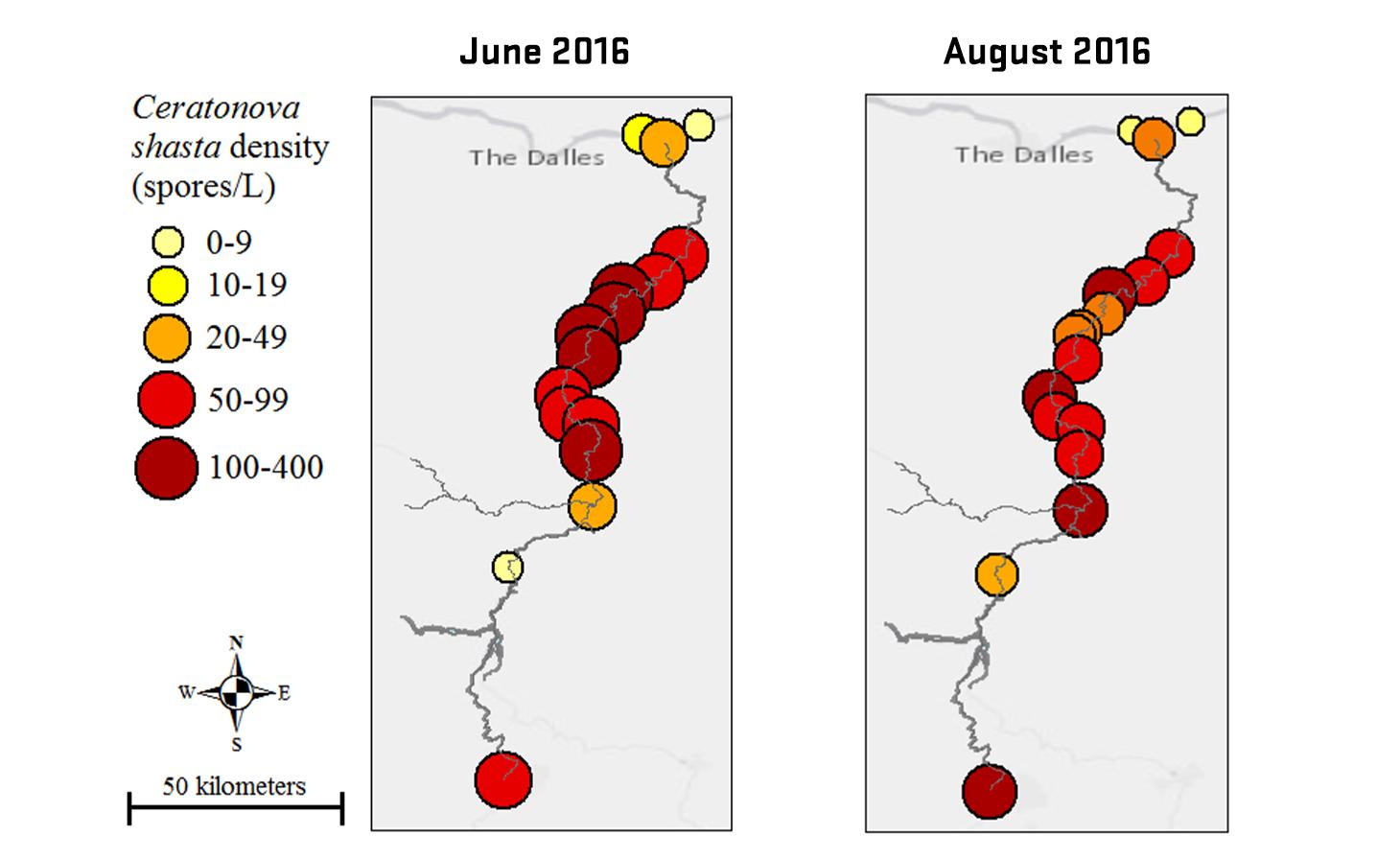
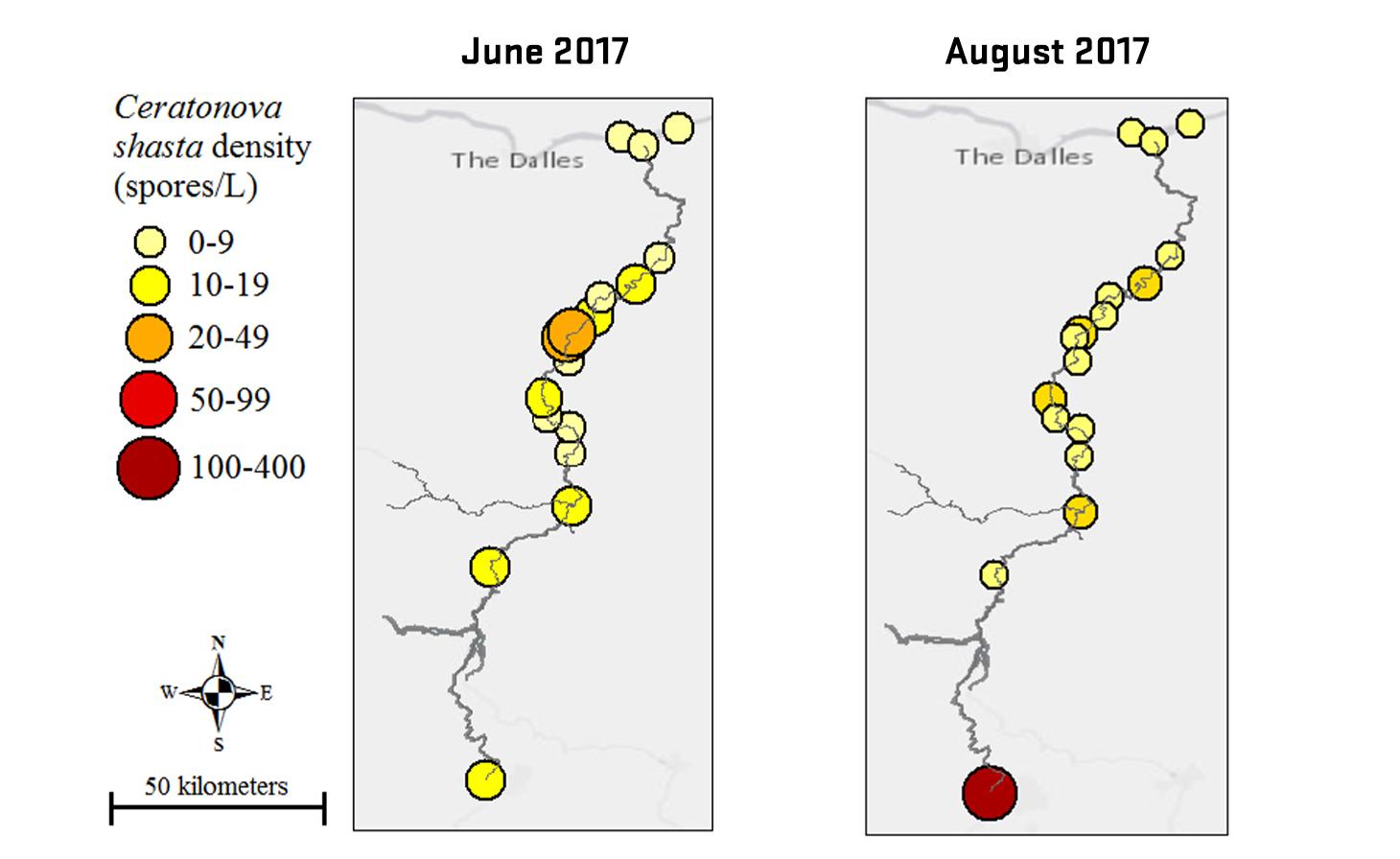
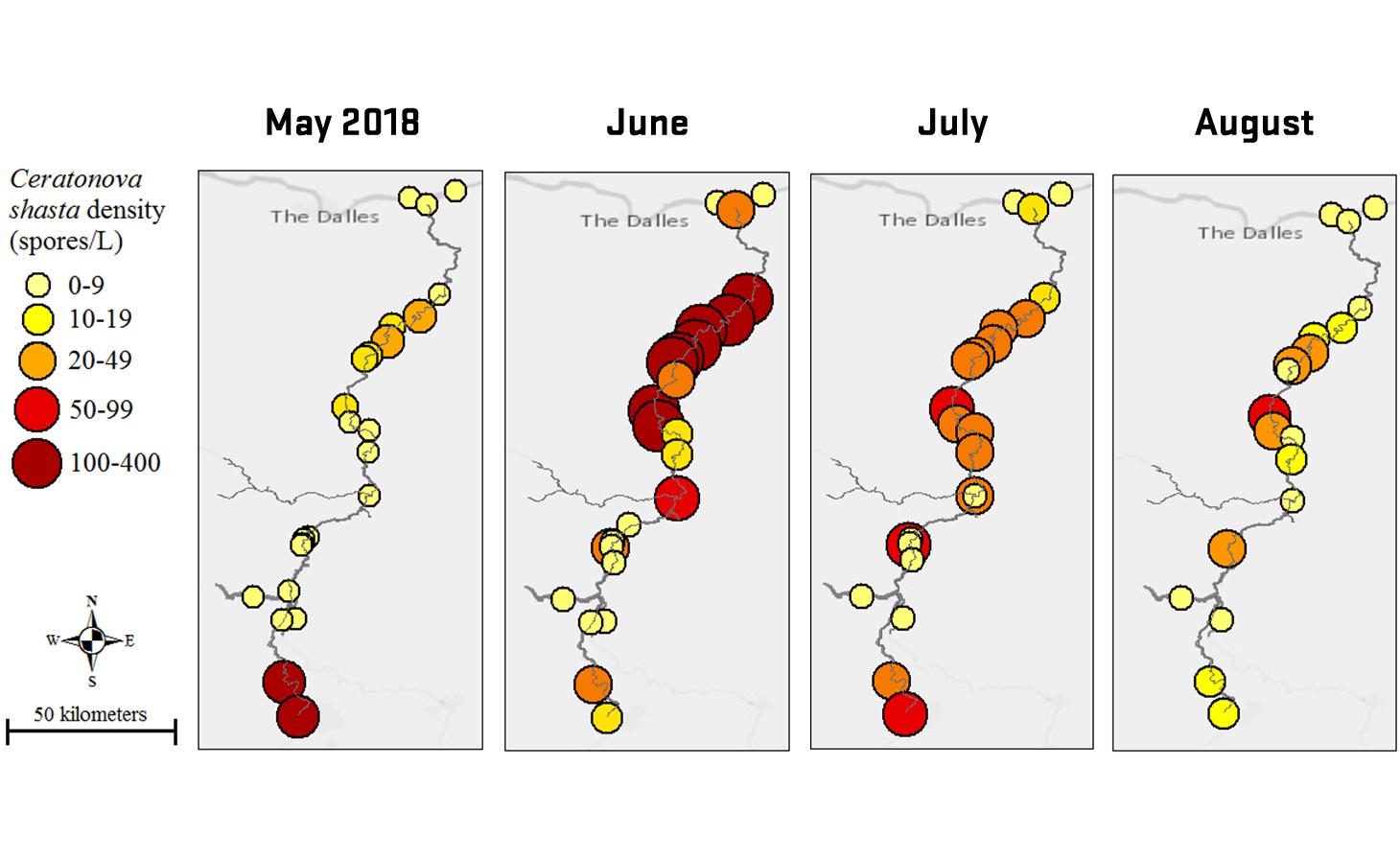
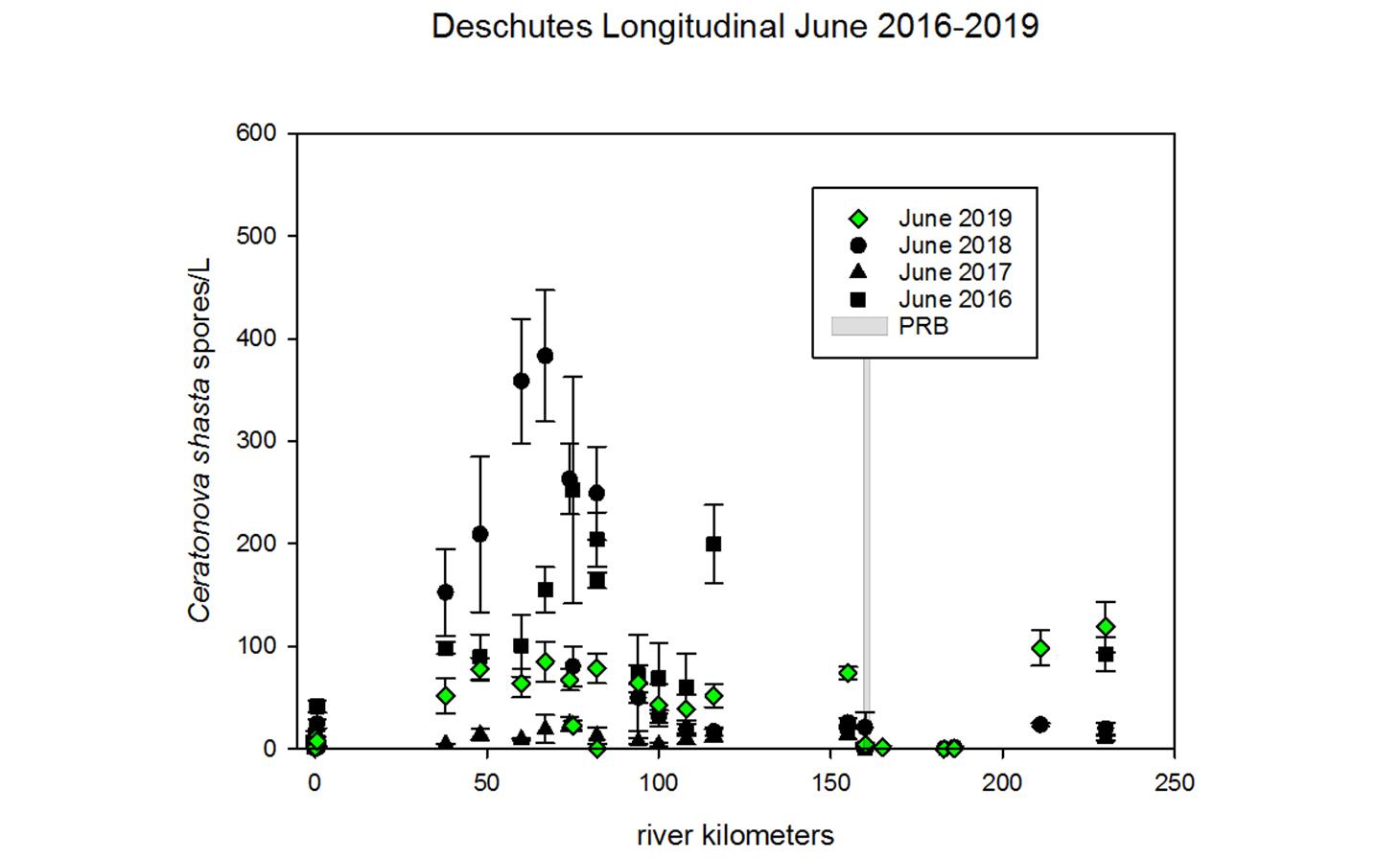
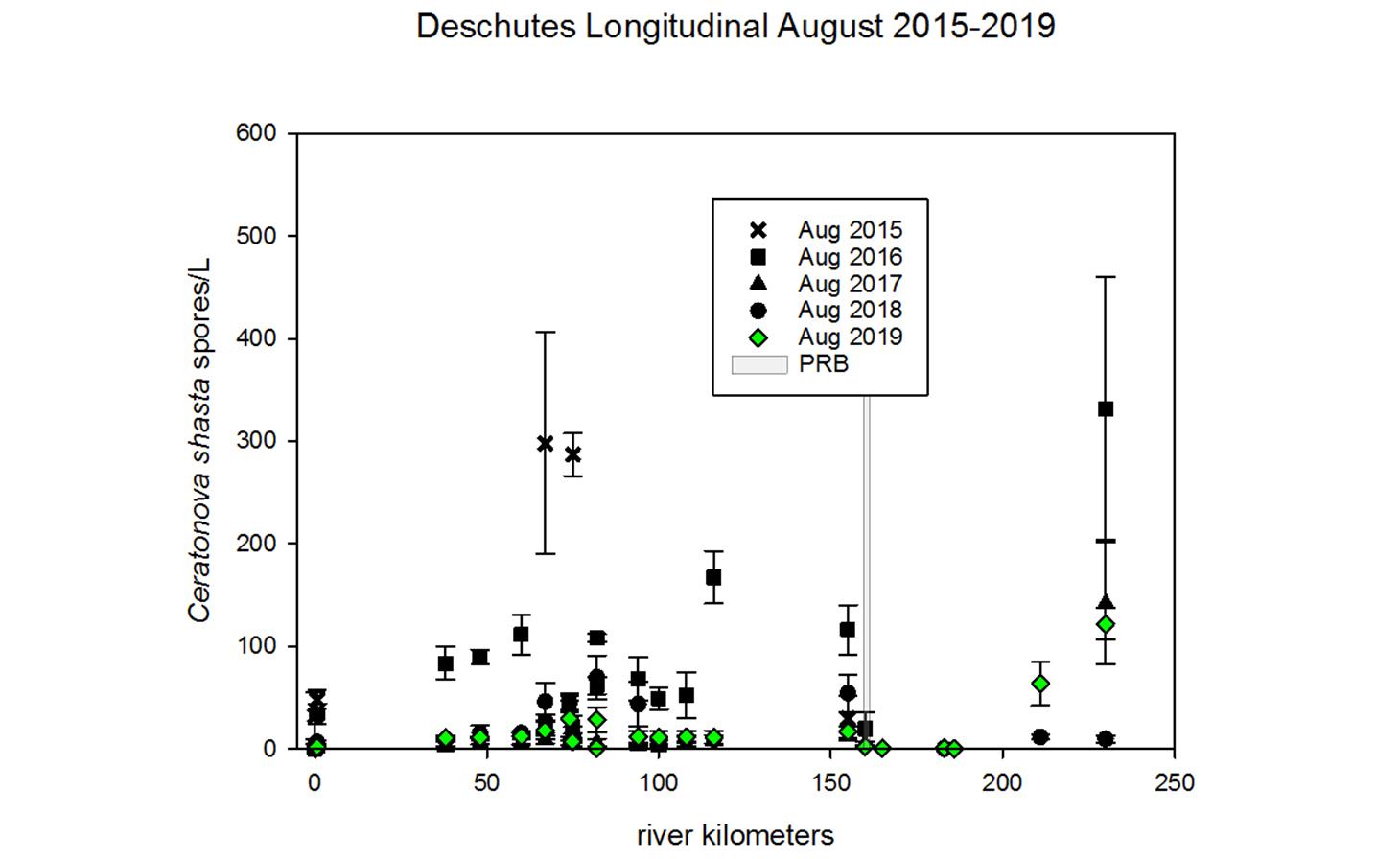
Through collaboration with the Confederated Tribes of the Warm Springs Reservation of Oregon (CTWSRO), USFWS, and ODFW we have developed a parasite monitoring program which contributes to our understanding of the role of pathogens (such as Ceratonova shasta) in juvenile and pre-spawn mortalities of adult Spring Chinook salmon of the Deschutes River Basin. Monitoring is primarily through quantification of waterborne stages of the parasite.