Projects
Projects
Our strategy
The Bermuda Institute for Ocean Sciences Collaboration for Ocean Processes and Ecology (BIOS-SCOPE)
Funded by Simons Foundation International
The Sargasso Sea is a natural laboratory for studying how ocean warming influences the movement of carbon from the atmosphere to the deep sea. Here, the ocean rhythmically transitions between cool, productive winter periods when nutrients are mixed to the surface, and warm, summer periods when nutrients are scarce and chlorophyll declines to extreme lows. These seasonal oscillations, which are normal in the northwestern Sargasso Sea, help us understand how ocean biology is changing with the global expansion of thermally stratified oceans.

BIOS-SCOPE researchers Craig Carlson (co-PI, UCSB) and Chris Suffridge (postdoc, OSU) deploy an in-situ seawater sampling system on a cruise in July, 2018.
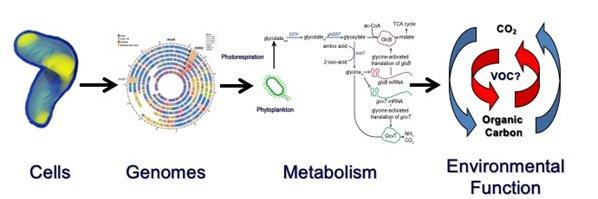
BIOS-SCOPE is an interdisciplinary scientific study of microbial carbon cycling in the northwestern Sargasso Sea that is funded by Simons Foundation International (SFI). With BIOS-SCOPE support, the Giovannoni lab has investigated genomes from SAR202 bacteria (Landry et al., 2017). These organisms, which we discovered in the 1990's, diversified early in bacterial evolution. By studying genome sequences, we were able to reconstruct hypothetical metabolic pathways that we propose evolved in ancient oceans for breaking down and oxidizing recalcitrant carbon molecules. With our BIOS-SCOPE partners, we are currently involved in laboratory and field research testing SAR202’s ability to metabolize recalcitrant carbon sources, thus elucidating the final stages of dissolved organic carbon oxidation by planktonic microbes.
An additional BIOS-SCOPE funded investigation applies network theory to long-term microbial community structure data from the Bermuda Atlantic Time Series (BATS) Program to demonstrate that plankton community structure is primarily determined by the environment. While this is mostly true, in some parts of the plankton Tree-of-Life, Hubble's neutral theory may apply, meaning that bacteria can sometimes substitute for evolutionary neighbors in food webs (Vergin et al., 2017).
The North Atlantic Aerosols and Marine Ecosystems Study
Funded by NASA grant no. NNX15AE70G
Seasonal phytoplankton blooms in the North Atlantic are one of Earth’s largest annual biological events. The NASA funded NAAMES Project focuses particularly on interactions between plankton blooms and the atmosphere. Our role in NAAMES is to measure plankton diversity by sequencing rRNA genes, and use the resulting data to reconstruct the complex biology of blooms. As part of NAAMES, we also work with OSU faculty member Dr. Kim Halsey to study the cycling of volatile organic compounds (VOCs) by plankton cells. Dr. Halsey's team focuses on VOC production by phytoplankton, whereas we use the HTCL's unique capabilities to study VOC oxidation.

The NAAMES science crew on the North Atlantic, in Dec. 2015.
The study of VOCs both in lab and in the environment is enabled by our proton transfer reaction time of flight mass spectrometer (PTR-MS), which was purchased by the NAAMES grant. This sensitive and specialized instrument allowed us to discover new pathways for the production of dimethylsulfide (DMS) in SAR11 bacteria (Sun et al., 2016), and recently we used the PTR-MS it to uncover new pathways of arsenic cycling by plankton (Giovannoni et al., 2019).
Dissolved Organic Carbon Cycling by SAR11 Marine Bacteria
Funded by National Science Foundation Grant OCE-1436865 and the U.S. Department of Energy Joint Genome Institute under Contract No. DE-AC02-05CH11231.
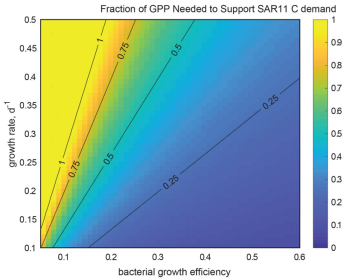
The fraction of Gross Primary Production needed to support SAR11 carbon demand over a range of assumed bacterial growth efficiencies and specific growth rates. The color axis is fixed from 0-1. (White et al., 2019)
This project is focused on SAR11, the most abundant group of bacterioplankton in the oceans. Globally, they are estimated to oxidize between 6 and 37% of all the organic carbon (gross primary production; GPP) produced by photosynthesis each day in the oceans (White et al, 2019). The activities of bacterioplankton determine the oceanic residence times of different forms of organic carbon, and ultimately shape the composition of the marine dissolved organic carbon pool, which nearly equals atmospheric CO2 in total mass. We culture cells and sequence genomes, and then analyze that data to predict carbon oxidation biochemistry. We experiment with cells in culture to test genome-based hypotheses, and then use research ships to investigate the importance of the new discoveries in an ecological context. Accurate and detailed information provided by our research is used in models that are valued for their potential to predict and understand future changes in ocean ecosystems.
In this research we partner with the Trent Northen of the Joint Genome Institute, applying metabolomics by mass spectrometry to study SAR11 interactions with DOM. Our goal in this research is to identify new compounds metabolized by SAR11 and variation in carbon metabolism between different SAR11 ecotypes.
Unraveling Thiamin Cycling Complexity and Impacts on Microbial Networks
Funded by National Science Foundation Grant DEB-1639033
Thiamin (vitamin B1) is an essential coenzyme across all domains of life that catalyzes key steps in carbon metabolism. Interestingly, many marine phytoplankton and bacteria have lost the ability to synthesize vitamin B1, and must rely on dissolved B1 to meet their metabolic demands. Recently it has been shown that obligate requirement for vitamin B1 (auxotrophy), takes many forms, with different groups of organisms lacking different portions of the thiamin biosynthetic pathway (Carini et al., 2014). We are investigating thiamin metabolism by plankton cells, and are measuring the concentrations of thiamin, its biosynthetic precursors, and its degradation products in seawater. Our goal is to better understand how thiamin metabolism evolved, and whether the availability of thiamin-compounds controls blooms of plankton in the oceans. Our collaborator on this project is Prof. Dr. Alexandra Z. Worden of GEOMAR, the Helmholtz Centre for Ocean Research in Kiel.
Regulation of Nutrient Assimilation in Streamlined Oligotrophic Microorganisms
Funded by National Science Foundation NSF 183844
Many microorganisms are adapted to life at very low nutrient concentrations. Some microbes that grow at these low nutrient concentrations – oligotrophs - do not regulate biochemical pathways for nutrient assimilation involving carbon oxidation. In contrast, among the more commonly studied copiotrophs - cells that grow rapidly at high nutrient concentrations - regulation of nutrient assimilation is ubiquitous. An example of a cell that has reduced regulation is Pelagibacter (SAR11), the most abundant organism in the oceans. At present there is no explanation for the lack of regulatory systems for nutrient uptake in cells that live in low nutrient ecosystems.
In this study, a combination of laboratory experiments, mathematical modeling and genome analysis is being used to examine hypotheses about the costs and benefits of regulating nutrient assimilation. The mathematical modeling component of this project is a partnership with Ferdi Hellweger of Berlin Technical University. Studying the costs and benefits of different strategies for nutrient assimilation in microbes will aid in the prediction of behavior of nutrient cycles and will lead to a better understanding of aquatic ecosystems. This research may also help us understand why so many common microbial cell types have been challenging to grow and study in scientific laboratories.
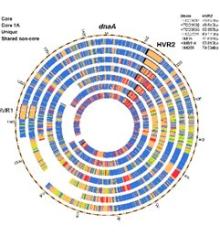
Mechanisms That Produce Pan-Genome Diversity in Globally Abundant Bacteria
Joint Genome Institute User grant, with Co-I.'s T. Sharpton and B. Temperton
Bacteria are dazzling in their ability to adapt by acquiring foreign genetic material, a process known as horizontal gene transfer (HGT). Most of this genetic material is concentrated in hypervariable genome regions (HVR’s), which play an important role in adaptive evolution. Little is known about the origins and variability of this rapidly changing genetic material. This project develops new metagenomics approaches for studying HVR variation and evolution. It addresses the broad question of how bacteria adapt to change by focusing on the specific example of HVR2 in Pelagibacterales, which are the most abundant microorganisms on the planet. Examining relationships between natural variation in HVRs and typical estimators of population diversity will clarify how microbial life adapts to environmental change on short time scales. These issues are fundamental to understanding how bacteria evolve.